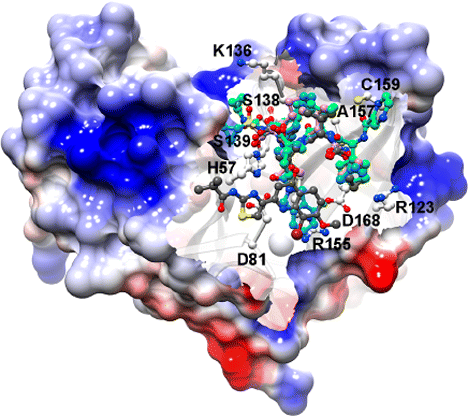
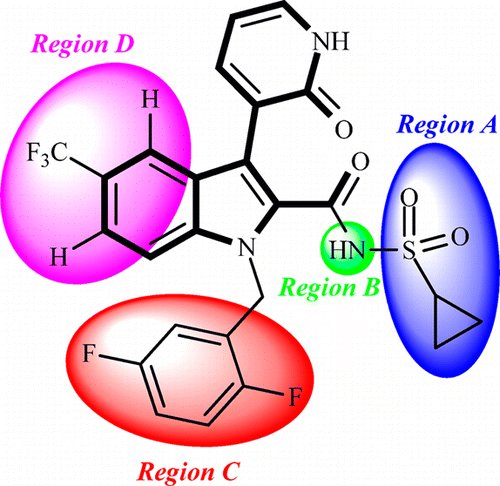
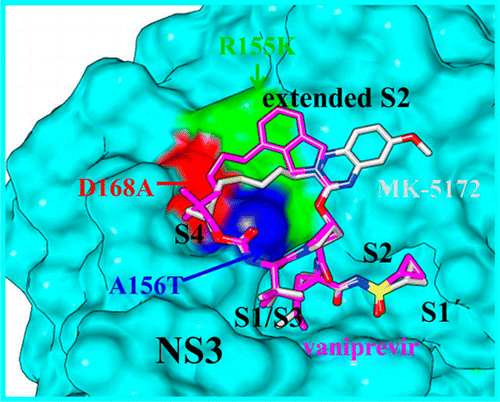
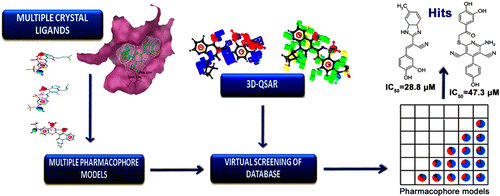
The Nobel Prize in Physiology or Medicine 2020 was awarded to Harvey J. Alter, Michael Houghton, and Charles M. Rice for their discovery of the hepatitis C virus. Their work paved the way for decades of important research, leading to sensitive blood tests for the disease, as well as antiviral drugs that can cure it. […]

The Nobel Prize in Physiology or Medicine 2020 was awarded to Harvey J. Alter, Michael Houghton, and Charles M. Rice for their discovery of the hepatitis C virus. Their work paved the way for decades of important research, leading to sensitive blood tests for the disease, as well as antiviral drugs that can cure it.
While these breakthroughs have created the possibility of someday eliminating the disease, for now it still presents a major global health risk. Researchers around the world continue to study hepatitis C and look for new and more effective ways of detecting it, stopping its spread, and treating patients who contract it.
In honor of the achievements of this years’ Nobel laureates, ACS Publications presents a collection of recent important hepatitis C research from the Journal of Medicinal Chemistry and ACS Infectious Diseases.
***
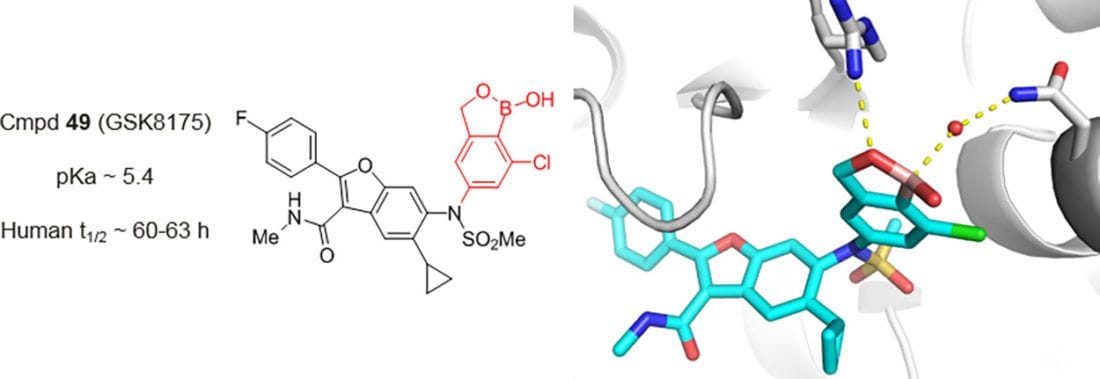
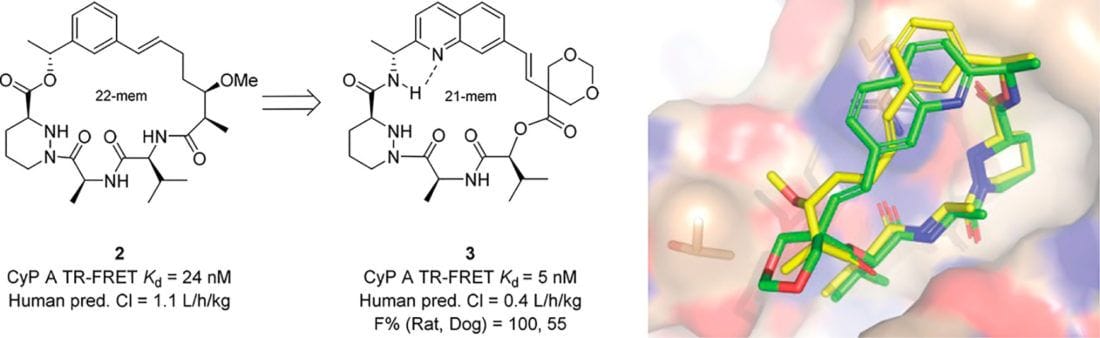
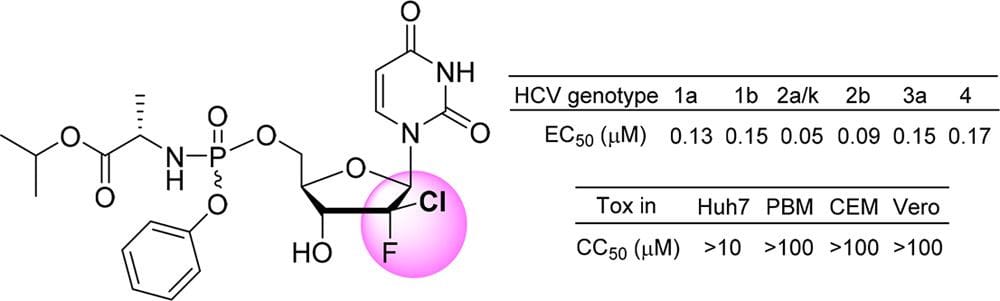
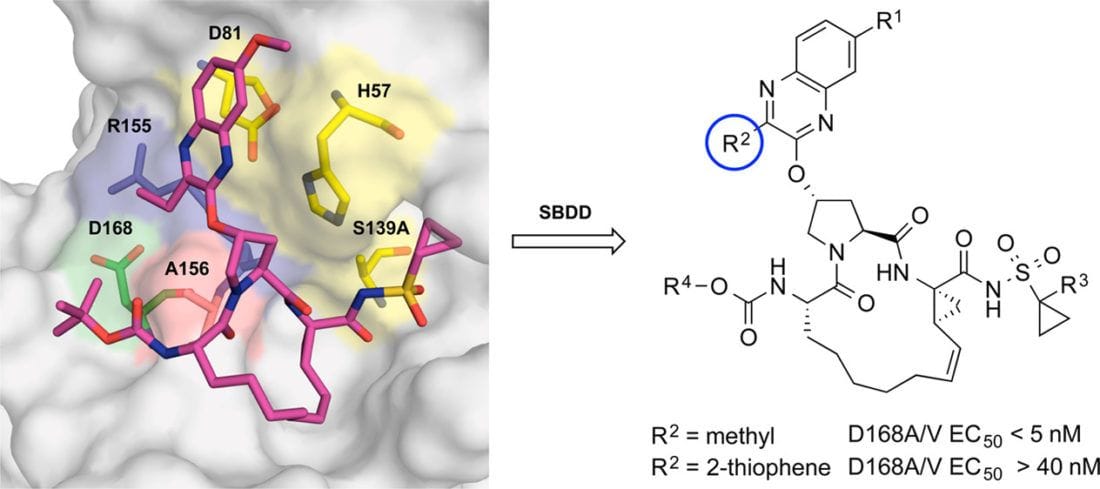
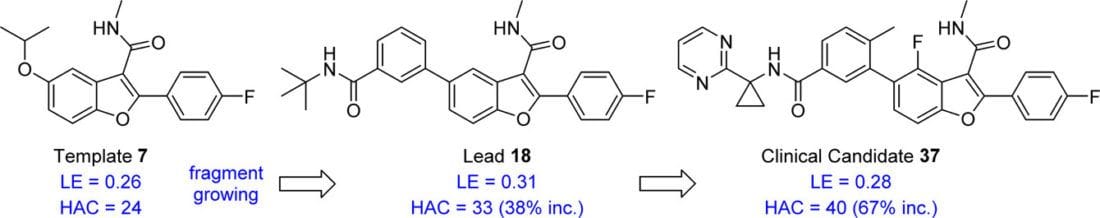
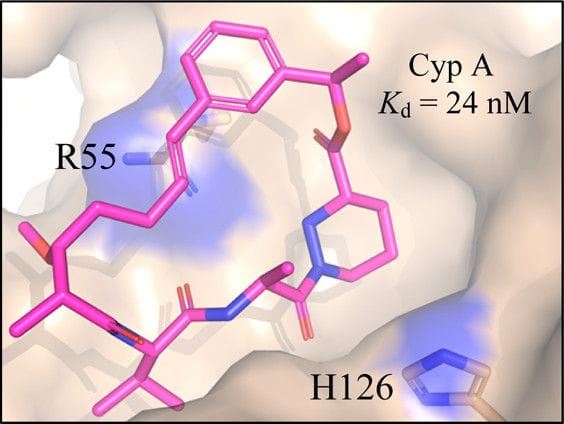
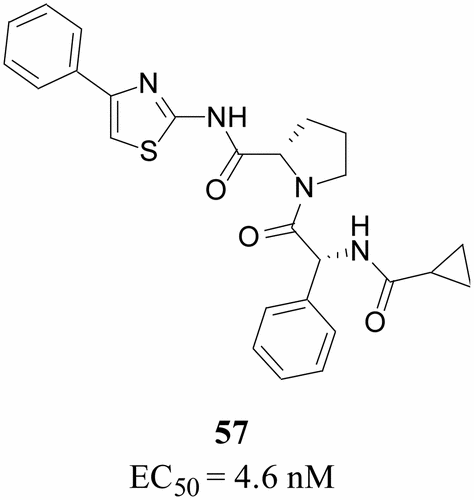
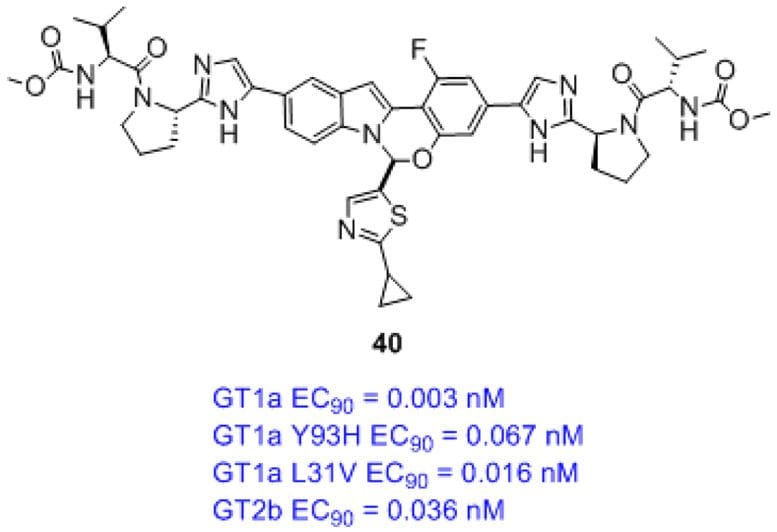
Design of N-Benzoxaborole Benzofuran GSK8175—Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor